OPFVTA Reexecution Study


This repository contains code, data, and prose as required to:
- re-execute analysis and production of the OPFVTA article in a container environment.
- generate a meta-article, detailing the reexecution environment and including a reference example of how to inspect reproducibility across reexecutions.
How to clone this repository
This repository contains submodules/subdatasets, which should be installed alongside the parent repository:
datalad install -r https://gin.g-node.org/TheChymera/opfvta-reexecution.git
cd opfvta-replication-2023
How to re-run
This reexecution system contains two targets, the original article, and publishing documents for the “meta-article”.
The latter can be executed in absence of the former, as it draws on the cummulative reexecution record found in the outputs/ directory.
Executing the former will produce your own reexecution file, which you can contribute to the record, and which all publishing documents created on your system will take into account.
For a graphical break-down, see the following workflow figure:
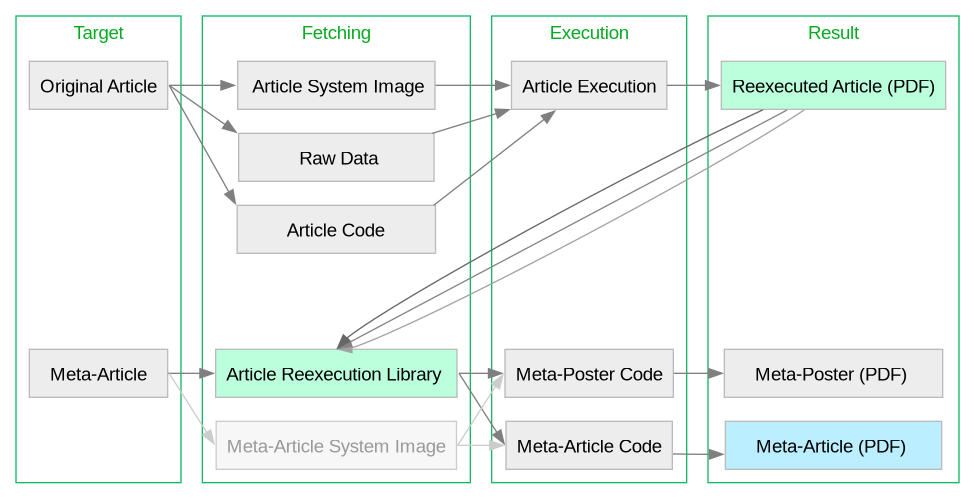
I. Reexecuting the OPFVTA Article
Warnings:
- We estimate that the analysis required more than 500GB, 400GB of which will be stored in a scratch directory, which is
./scratch/ by default and can be configured with the SCRATCH_PATH variable.
- The analysis self-limits RAM to run on less powerful systems
- Reexecuting the computation as well as the article is time consuming and resource-intensive, it is recommended to use a tool such as
tmux or screen to preserve long running processes.
First, retrieve the data and other large files:
make submodule-data
Note that the outputs/ directory will have some content from other
executions (organized by datetime) for comparison.
Once the required content has been fetched, you can reexecute the OPFVTA article via singularity or oci containers.
This step generates intermediate results in the scratch directory which are not preserved in this repository by default, as configured in scratch/.gitignore.
The final result is a PDF article and its associated elements (mainly volumetric binary data, .nii.gz files) which will be stored in a datestamped and annotated directory under outputs/.
Most large files, including the results are stored and versioned via git-annex and therefore present in this repository.
After executing the analysis the output will have a new entry for the data produced, which can be saved and recorded with datalad save.
For apptainer/singularity:
make analysis-singularity
or
With docker or podman, you can execute the analysis inside an OCI container.
make analysis-oci
The results of the analysis will be stored under outputs/<execution-environment>_<datestamp>
II. Reexecuting the Meta-Article
To avoid confusion, we use the term 'article' to refer to a version of the OPFVTA article, and 'meta-article' to refer to the paper regarding the reexecution process and findings.
Generation of the meta-article uses files generated by the OPFVTA analysis which are expected to be in the outputs/ directory.
Prior to generating the meta-article, outputs/ must contain the data from previous analyses, which is not locally available by default.
Note: Regenerating the OPFVTA article will create an additional pdf, but the previous pdfs are required to compare.
To fetch the OPFVTA analysis outputs:
datalad get outputs/*/article.pdf
Finally we generate new graphical elements and compile the text via LaTeX into a novel meta-article PDF.
The meta-article can then be generated by a container with all of the dependencies preinstalled using:
make container-article
or
If you prefer to run the generation outside of a container, you will need to install dependencies (suggested to use distribution package manager, packages below are debian names):
- laTex
- biber
- datalad
- diff-pdf
- graphviz
- matplotlib
- pandas
- seaborn
- sklearn
- statsmodels
- yaml
You will also need to install sourceserifpro font using the tlmgr.
make article
In either of those cases you can then view the produced meta-article at TODO.
Cleaning up between runs
The steps are designed to be idempotent, and some dynamically generated components will not be regenerated for subsequent runs.
If you are not merely trying to get a PDF to read or working on the human-readable text — but instead working on the figure-generating code — it is advisable to always deep-clean the dynamic elements in between re-making the article.
make article-clean && make article
Internal
We openly share all code and data via the Gin repository referenced above.
This open infrastructure is however slow, which may be particularly inconvenient for prolonged development work.
Trusted collaborators may instead prefer to use the smaug.dartmouth.edu remote.
To use this remote you should:
- Make sure you have SSH access to
smaug.dartmouth.edu, and have configured the host via your config file; you can do so by running:
cat >> ~/.ssh/config<< EOF
Host smaug
Hostname smaug.dartmouth.edu
AddKeysToAgent yes
Port <secret_port>
User <your_username_for_which_smaug_has_your_SSH_public_key>
EOF
- Add the remote to the Git repository, and make sure you are synced up:
cd path/to/your/repo
git remote add smaug smaug:/mnt/btrfs/datasets/incoming/con/opfvta-replication-2023.git
datalad get . -s smaug
- Optionally, and only if you are an internal contributor looking to push t, it makes sense to to automatically keep our three remotes (gin, smaug, GitHub) in sync, you should make use
